About
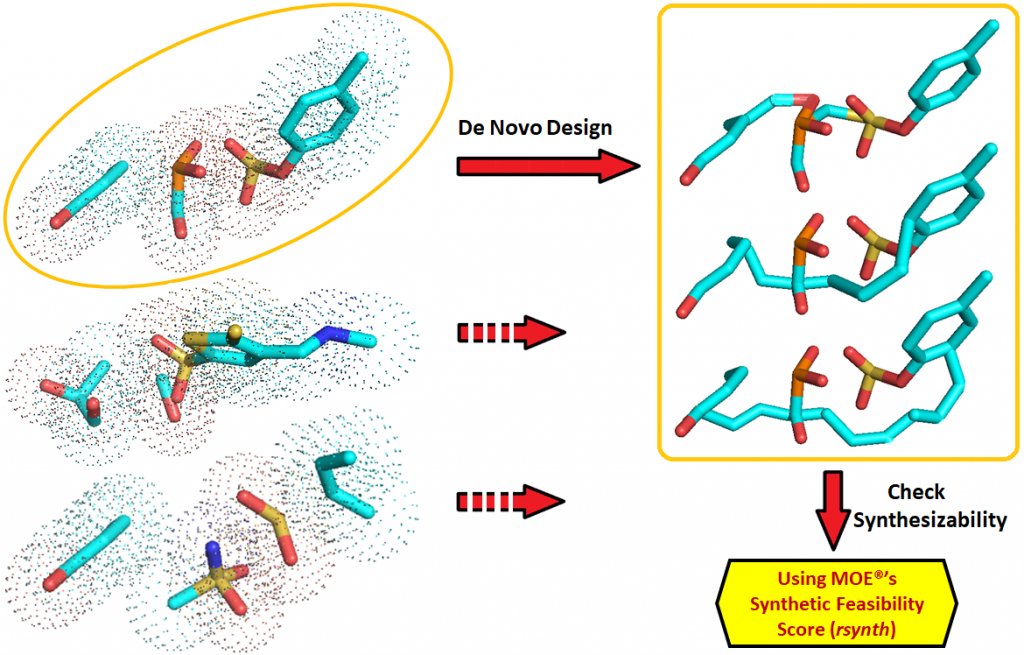
This method (FragDeNovo) utilizes artificial intelligence strategies (AI), such as data mining and machine learning algorithms, to support fragment-based de novo drug design. Having the binding pocket for a protein target of interest, the method will search for similar pockets in a database of native protein-ligand complexes. The search is based on 3D-geometric and chemical similarity of the atoms forming the binding pocket. For each similar pocket identified, the ligand’s sub-structures (fragments) corresponding to that pocket are extracted, thus, forming a virtual library of fragments. The fragments are used along with machine learning algorithms and specialized de novo drug design platform to create larger molecules that contain these fragments and are still predicted to bind. The method relies on efficient algorithms to facilitate the pocket similarity search and de novo drug design protocols, and it can be used for structure-based drug design. It is a breakthrough method in that it is using machine learning to engineer new molecular entities in ways that are different that conventional approaches.
The program to generate the virtual library of fragments (FragVLib) is Open Source software that is available at no charge. It is supplied as is, without warranty, and may be freely used and distributed under the terms of the Gnu General Public License (GPL).
To download the program, which runs under Linux, follow this link: FragDeNovo_to_public.tar.gz