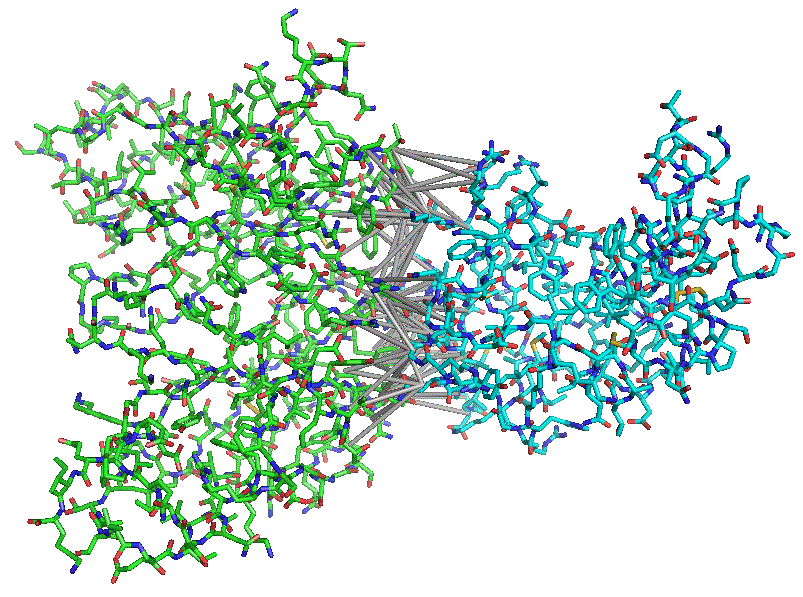
SPIDER (Scoring Protein Interaction Decoys using Exposed Residues) is developed as a knowledge-based scoring function for protein-protein interaction decoys. SPIDER is a novel multi-body pose-scoring function that has no theoretical limit on the number of residues contributing to the individual interaction terms. SPIDER’s score relies on the geometric similarity of interfacial residues between docking poses and naturally occurring (native) poses.
We use a coarse-grain representation of a protein-protein complex where each residue is represented by its side chain centroid. We apply a computational geometry approach called Almost-Delaunay tessellation that transforms protein-protein complexes into a residue contact network, or an un-directional graph where vertex-residues are nodes connected by edges. This treatment forms a family of interfacial graphs representing a dataset of protein-protein complexes. We then employ frequent subgraph mining approach to identify common interfacial residue patterns that appear in at least a subset of native protein-protein interfaces. The geometrical parameters and frequency of occurrence of each “native” pattern in the training set are used to develop the new SPIDER scoring function.
We demonstrated that SPIDER scoring function ranks native and native-like poses above geometrical decoys; SPIDER was ranked among the top 6 (out of 28) scoring functions in a recent round 21 of CAPRI (Critical Assessment of PRedicted Interactions) blind test of protein-protein docking methods (See References).
The program to calculate the SPIDER scores is Open Source software that is available at no charge. It is supplied as is, without warranty, and may be freely used and distributed under the terms of the Gnu General Public License (GPL).
To download the program, which runs under Linux (64-bit), follow this link: SPIDER_WORK.tar.gz
Raed Khashan, Weifan Zheng, and Alexander Tropsha. Scoring protein interaction decoys using exposed residues (SPIDER): A novel multibody interaction scoring function based on frequent geometric patterns of interfacial residues. Proteins: Structure, Function, and Bioinformatics, Volume 80, Issue 9, Pages 2207-2217, August 2012.
References
1. Raed Khashan, Weifan Zheng, and Alexander Tropsha. Scoring protein interaction decoys using exposed residues (SPIDER): A novel multibody interaction scoring function based on frequent geometric patterns of interfacial residues. Proteins: Structure, Function, and Bioinformatics, Volume 80, Issue 9, Pages 2207-2217, August 2012.
2. Sarel J. Fleishman, Timothy A. Whitehead, Eva-Maria Strauch, Jacob E. Corn, Raed Khashan, Stephen Bush, Denis Fouches, Alexander Tropsha, et al. Community-Wide Assessment of Protein-Interface Modeling Suggests Improvements to Design Methodology. Journal of Molecular Biology, Volume 414, Issue 2, Pages 289-302, November 2011.